|
|
4 жил өмнө | |
|---|---|---|
| .. | ||
| README.md | 4 жил өмнө | |
| arguments.py | 4 жил өмнө | |
| requirements.txt | 4 жил өмнө | |
| run_trainer.py | 4 жил өмнө | |
| run_training_monitor.py | 4 жил өмнө | |
| tokenize_wikitext103.py | 4 жил өмнө | |
| utils.py | 4 жил өмнө | |
README.md
Training ALBERT with decentralized averaging
This tutorial will walk you through the steps to set up collaborative training with the ALBERT-large-v2 model and the WikiText103 dataset. It uses huggingface datasets and transformers libraries to compute local updates, using hivemind.CollaborativeOptimizer to exchange information between peers.
Preparation
- Install hivemind:
pip install git+https://github.com/learning-at-home/hivemind.git - Dependencies:
pip install -r requirements.txt - Preprocess data:
python tokenize_wikitext103.py - Upload an archive preprocessed data to somewhere volunteers can reach, example:
https://hivemind-data.s3.us-east-2.amazonaws.com/wikitext103_preprocessed.tar
Running an experiment
Run the first DHT peer to welcome trainers and record training statistics (e.g. loss, performance):
- In this example, we use wandb.ai to plot training metrics; If you're unfamiliar with Weights & Biases, here's a quickstart tutorial.
- Run
python run_training_monitor.py --experiment_prefix NAME_YOUR_EXPERIMENT --wandb_project WANDB_PROJECT_HERE NAME_YOUR_EXPERIMENTmust be a unique name of this training run, e.g.my-first-albert. It cannot contain.due to naming conventions.WANDB_PROJECT_HEREis a name of wandb project used to track training metrics. Multiple experiments can have the same project name.$ python run_training_monitor.py --experiment_prefix my-albert-v1 --wandb_project Demo-run [2021/06/17 16:26:36.083][INFO][root.log_visible_maddrs:42] Running a DHT peer. To connect other peers to this one, use --initial_peers /ip4/8.8.8.8/tcp/1337/p2p/XXXX /ip4/8.8.8.8/udp/31337/quic/p2p/XXXX wandb: Currently logged in as: XXX (use `wandb login --relogin` to force relogin) wandb: Tracking run with wandb version 0.10.32 wandb: Syncing run dry-mountain-2 wandb: View project at https://wandb.ai/XXX/Demo-run wandb: View run at https://wandb.ai/XXX/Demo-run/runs/YYY wandb: Run data is saved locally in /path/to/run/data wandb: Run `wandb offline` to turn off syncing. [2021/04/19 02:26:41.064][INFO][optim.collaborative.fetch_collaboration_state:323] Found no active peers: None [2021/04/19 02:26:44.068][INFO][optim.collaborative.fetch_collaboration_state:323] Found no active peers: None ... [2021/04/19 02:37:37.246][INFO][__main__.<module>:194] Step #1 loss = 11.05164 [2021/04/19 02:39:37.441][INFO][__main__.<module>:194] Step #2 loss = 11.03771 [2021/04/19 02:40:37.541][INFO][__main__.<module>:194] Step #3 loss = 11.02886
To join a collaboration with a GPU trainer,
- install the same dependencies (minus the
wandbandwhatsmyip), download the data and unpack it to the experiment folder, - if necessary, specify paths:
--dataset_path ./path/to/unpacked/data --tokenizer ./path/to/tokenizer/config(see default paths for reference) run:
python run_trainer.py \ --experiment_prefix SAME_AS_IN_RUN_TRAINING_MONITOR --initial_peers ONE_OR_MORE_PEERS --seed 42 \ --logging_first_step --logging_steps 100 --output_dir ./outputs --overwrite_output_dir --logging_dir ./logs
Here,
ONE_OR_MORE_PEERSstands for multiaddresses of one or multiple existing peers (training monitors or existing trainers) collected from the first lines of their terminal output. For the example above, the multiaddresses would be:--initial_peers /ip4/8.8.8.8/tcp/1337/p2p/XXXX /ip4/8.8.8.8/udp/31337/quic/p2p/XXXXNote: a multiaddress is a format for encoding multiple layers of addressing information that supports a number of different protocols. In hivemind, we typically operate with multiaddresses that contain a libp2p peer ID (e.g.
/p2p/XXXX) together with the information about how to reach it (e.g. the IPv4 address and TCP port/ip4/8.8.8.8/tcp/31337or the information about a relay used for NAT traversal).You may need to change the IP address to a publicly visible one if some of the initial peers are located behind NAT. If you have any trouble doing this, consider the "Using IPFS" section.
- install the same dependencies (minus the
See the "Tips and tricks" section for more information on setting up collaborative training.
As the peer begins training, it will periodically report training logs in the following form:
[...][INFO][...] Collaboration accumulated 448 samples from 17 peers; ETA 18.88 seconds (refresh in 15.73s.)
[...][INFO][...] Collaboration accumulated 4096 samples from 16 peers; ETA 0.00 seconds (refresh in 0.50s.)
[...][INFO][optim.collaborative.step:195] Averaged tensors successfully with 17 peers
[...][INFO][optim.collaborative.step:211] Optimizer step: done!
06/17/2021 18:58:23 - INFO - __main__ - Step 0
06/17/2021 18:58:23 - INFO - __main__ - Your current contribution: 892 samples
06/17/2021 18:58:23 - INFO - __main__ - Local loss: 11.023
Sanity check: a healthy peer will periodically report Averaged tensors successfully with [N > 1] peers.
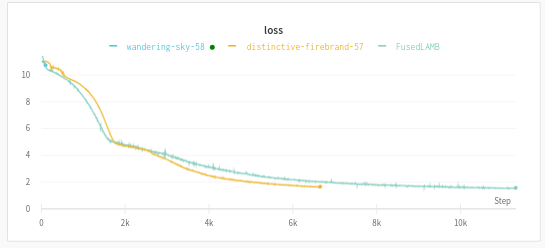
For convenience, you can view (and share!) the learning curves of your collaborative experiments in wandb:
Tips and tricks
Finally, we provide best practices for running collaborative experiments of different sizes.
Hosting the data
For small experiments (3-16 peers, <1GB data), you can use a free-tier file hosting that has a convenient way to [download with curl/wget](https://superuser.com/questions/470664/how-to-download-dropbox-files-using-wget-command). However, these services are not meant for high load and could ban you for generating too much traffic. If you want to scale up, you could either use an S3-like storage from [any](https://aws.amazon.com/s3/) [cloud](https://cloud.google.com/storage) [provider](https://cloud.google.com/storage) or host the data [yourself]((https://gist.github.com/willurd/5720255)). Large data files (>5GB) will take long to download; we recommend splitting them into chunks and implementing a custom dataloader that can load chunks on the fly. Finally, the most comme il faut solution to sharing large datasets is to use academic torrents.
run_training_monitor.py
This peer exists solely to welcome other peers onto the DHT and track learning progress. It requires neither GPU nor high bandwidth, the only prerequisite is that coordinator should have high uptime. If no high uptime server is available, one can also run multiple coordinators on different servers and list all of them as --initial_peers. The system will stay up as long as at least one coordinator is available. For short- to mid-term experiments you can host coordinator on a free-tier VM.
Tuning for hardware/network
The optimal training parameters for each peer depend on its GPU and internet connection. If a peer cannot accept incoming connections (e.g. when in colab or behind a firewall), add --client_mode to the training script (see example below). In case of high network latency, you may want to increase --averaging_expiration by a few seconds or set --batch_size_lead to start averaging a bit earlier than the rest of the collaboration. GPU-wise, each peer should be able to process one local microbatch each 0.5~1 seconds (see trainer's progress bar). To achieve that, we recommend tuning --per_device_train_batch_size and --gradient_accumulation_steps. The example trainer supports multiple GPUs via DataParallel. However, using advanced distributed training strategies (e.g. ZeRO-3) will require changes in run_trainer.py.
Using public GPU providers
There are awesome services like Google Colab, Kaggle kernels orPaperspace that provide free GPUs. These services usually come with significant limitations (e.g. last gen GPUs, reset every few hours), but they allow just about anyone to join your collaborative experiment. Here's how to best use them.
- before you begin, read the rules carefully. Most free-tier GPU services allow only one GPU per user and using more than one account will get you banned. It is your duty to make sure that collaborators won't get in trouble for helping you.
- most free GPUs are running behind a firewall, which requires you to run trainer with
--client_mode(see example below). Such peers can only exchange gradients if there is at least one non-client-mode peer (GPU server or desktop with public IP). We recommend using a few preemptible instances with the cheapest GPU you can find. For example, we tested this code on preemptibleg4dn.xlargenodes for around $0.15/h apiece with 8 AWS nodes and up to 61 Colab/Kaggle participants. - you can create starter notebooks to make it more convenient for collaborators to join your training run (example). Ideally, joining collaboration should take at most a couple of clicks.
Here's an example of a full trainer script for Google Colab:
!pip install transformers datasets sentencepiece torch_optimizer==0.1.0
!git clone https://github.com/learning-at-home/hivemind && cd hivemind && pip install -e .
!curl -L YOUR_HOSTED_DATA | tar xzf - # example: https://hivemind-data.s3.us-east-2.amazonaws.com/wikitext103.tar.gz
!ulimit -n 4096 && python ./hivemind/examples/albert/run_trainer.py \
--client_mode --initial_peers ONE_OR_MORE_PEERS --averaging_expiration 10 \
--batch_size_lead 300 --per_device_train_batch_size 4 --gradient_accumulation_steps 1 \
--logging_first_step --logging_steps 100 --output_dir ./outputs --overwrite_output_dir --logging_dir ./logs \
--experiment_prefix EXPERIMENT_NAME_HERE --seed 42
Using IPFS
If the initial peers for your experiment are located behind NAT and/or you have any trouble with figuring out their public IP addresses and ports, you can set up hivemind to use the IPFS network to find the route to your peers automatically. To do this, you should specify the --use_ipfs option on all peers (both training monitors and trainers) you are starting.
After that, it is enough to provide only a libp2p peer ID (e.g. /p2p/XXXX) for each initial peer. No other information (like IP addresses or TCP/UDP ports) is required.